Using The Generate Sequences Tool#
This tutorial teaches you how to generate functional sequences conditioned on the sequence context provided by a prompt. You will learn how to generate a sequence, then interpret and fine-tune the results. Use this as a starting point for generating a diverse library without existing experimental data.
If you run into any challenges or have questions while getting started, please contact OpenProtein.AI support.
What You Need Before Starting#
This tool requires a multiple sequence alignment (MSA), from which it builds a prompt. You can choose an existing prompt, upload your own MSA or have the OpenProtein.AI model generate one for you. If you aren’t already familiar with prompts, we recommend learning more about OpenProtein.AI’s prompts and prompt sampling methods before diving in.
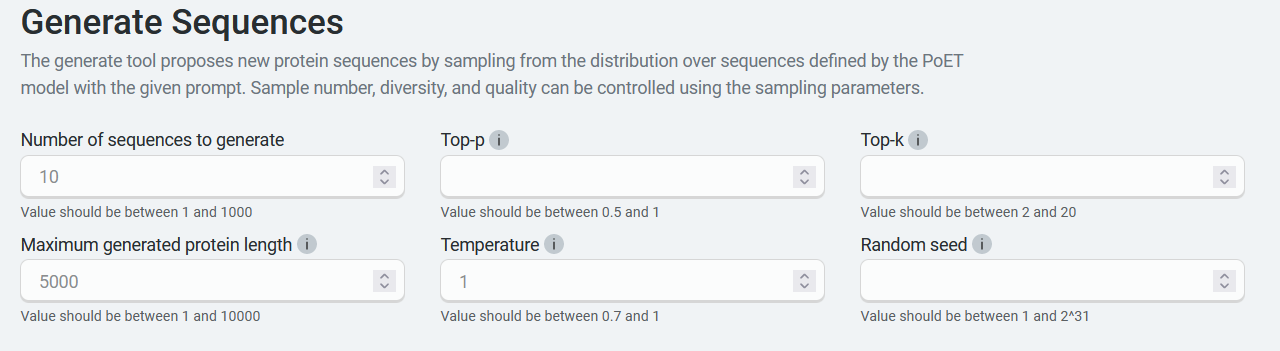
You also need to know about sampling parameters, which are settings that regulate randomness. These include temperature, top-p, and top-k.
Top-p (also known as nucleus sampling) limits sampling to amino acids with sum likelihoods which do not exceed the specified value. As a result, the list of possible amino acids is dynamically selected based on the sum of likelihood scores achieving the top-p value. For example, setting a top-p of 0.8 limits sampling to amino acids summing to an 80% or greater probability. Other amino acids are ignored.
Top-k limits sampling to a shortlist of amino acids, where the top-k parameter sets the size of the shortlist. For example, setting top-k to 5 means the model samples from the 5 likeliest amino acids at each position. Other amino acids are ignored.
Temperature is a number used to tune the degree of randomness. A lower temperature means less randomness; a temperature of 0 will always yield the same output.
A note on the Random seed setting: this determines the state of the random number generator for random sampling. If it is set to a specific number, the algorithm will sample the same set of sequences each time. We recommend not defining this seed unless you are reproducing a job.
Generating Sequences#
Navigate to the tool by opening the PoET dropdown menu, then selecting Generate Sequences. You can choose the model used to run the job. We recommend using PoET-2 for most use cases.
Step 1: Prompt Query#
Refer to Creating a Query to learn about Prompt Query.
Step 2: Prompt Context#
Refer to Creating a Context to learn about Prompt Context.
Step 3: Sampling Settings#
Set your parameters to control sampling behavior. In particular, temperature, top-p, and top-k provide the ability to focus sampling around highly likely sequences. We recommend that you use either top-p or top-k on a given job, not both. You can choose the default structure prediction model to generate the sequence structures after the job completes.
You’re ready to generate custom sequences! Click Run. The job may take a few minutes depending on how busy the service is, how long your sequences are, and how many sequences you want to score.
A 400 (Bad request) error code may be due to the following:
Issue description |
Solution |
|---|---|
Invalid PoET Job or Parent |
Re-enter prompt and try again. |
Invalid prompt in PoET service |
Reupload prompt and try again. Refer to the article about prompts. Ensure minimum and maximum similarity parameters are not filtering out all sequences in prompt. |
Invalid user input in align service |
Ensure you don’t have
If necessary, refer to the article on sampling parameters. |
Invalid MSA (not aligned, etc) |
|
Please contact OpenProtein.AI support if the suggested solutions don’t resolve the issue.
Interpreting Your Results#
Refer to Interpreting PoET Results Table.
Fine-tuning Your Results#
Improve your results by adding more sequences with your desired properties to your MSA, or by adjusting the prompt sampling method. You can also adjust the Maximum similarity to seed sequence and Minimum similarity to seed sequence fields.
If your results are too diverse, try adjusting temperature downwards to decrease the diversity of the sampling.
To improve scores, increase the number of the ensemble setting. This will result in higher scoring sequences, but will take longer to complete.
Next Steps#
Now that you can generate custom sequences, use the Structure Prediction tool on high scoring sequences to visualize their structural implication or use Substitution Analysis to view possible improvements to a sequence.