Aligning sequences#
Accessing the alignment tool#
To access the alignment tool, start by navigating to the global navigation located at the top right corner of the application. Select ‘Align Sequences’ to be redirected to the main alignment page, where you can perform sequence alignment tasks.
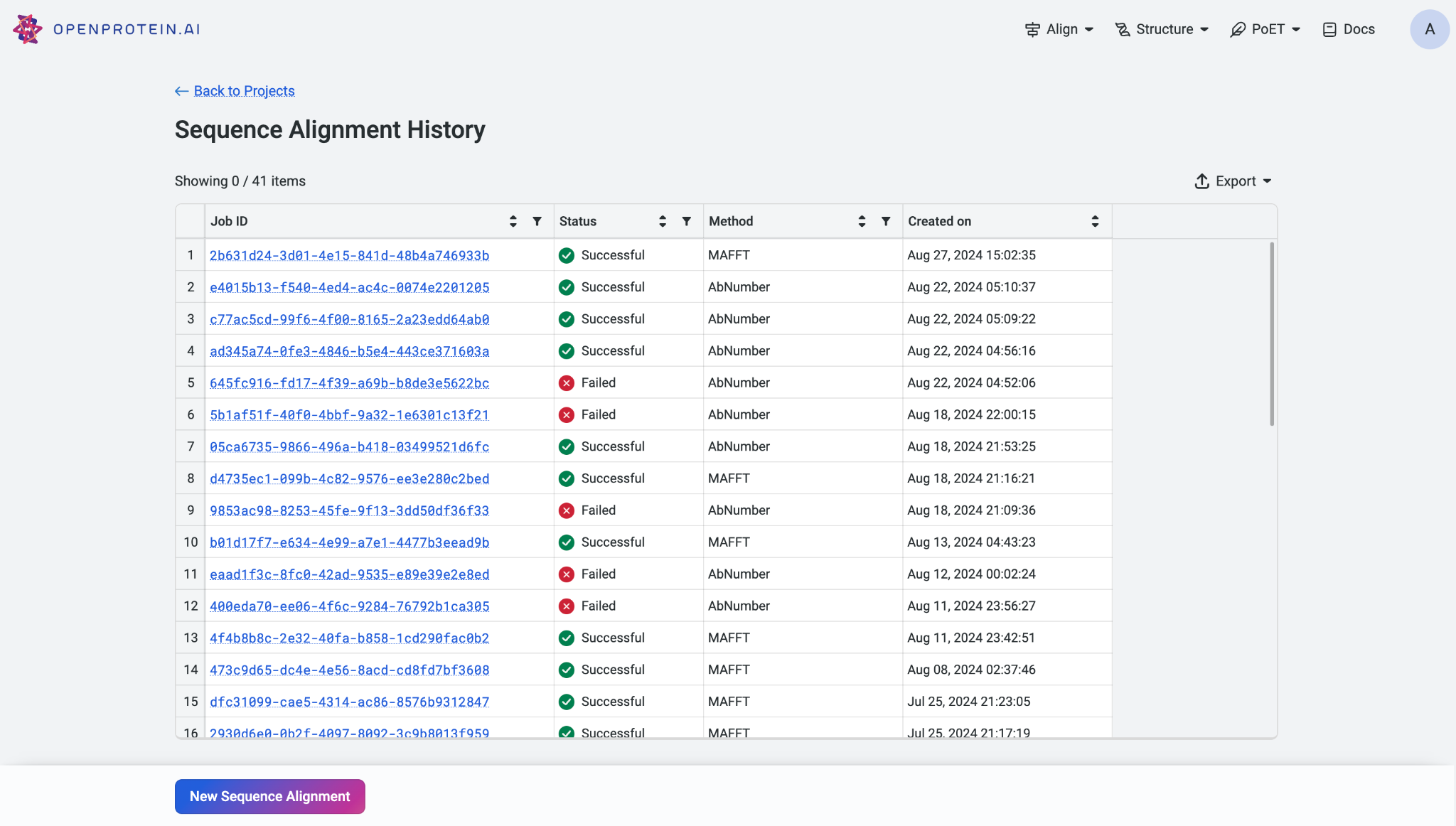
If you need to view past alignment jobs, select ‘History’ to access the history page displaying your previous alignments.
Uploading your sequences#
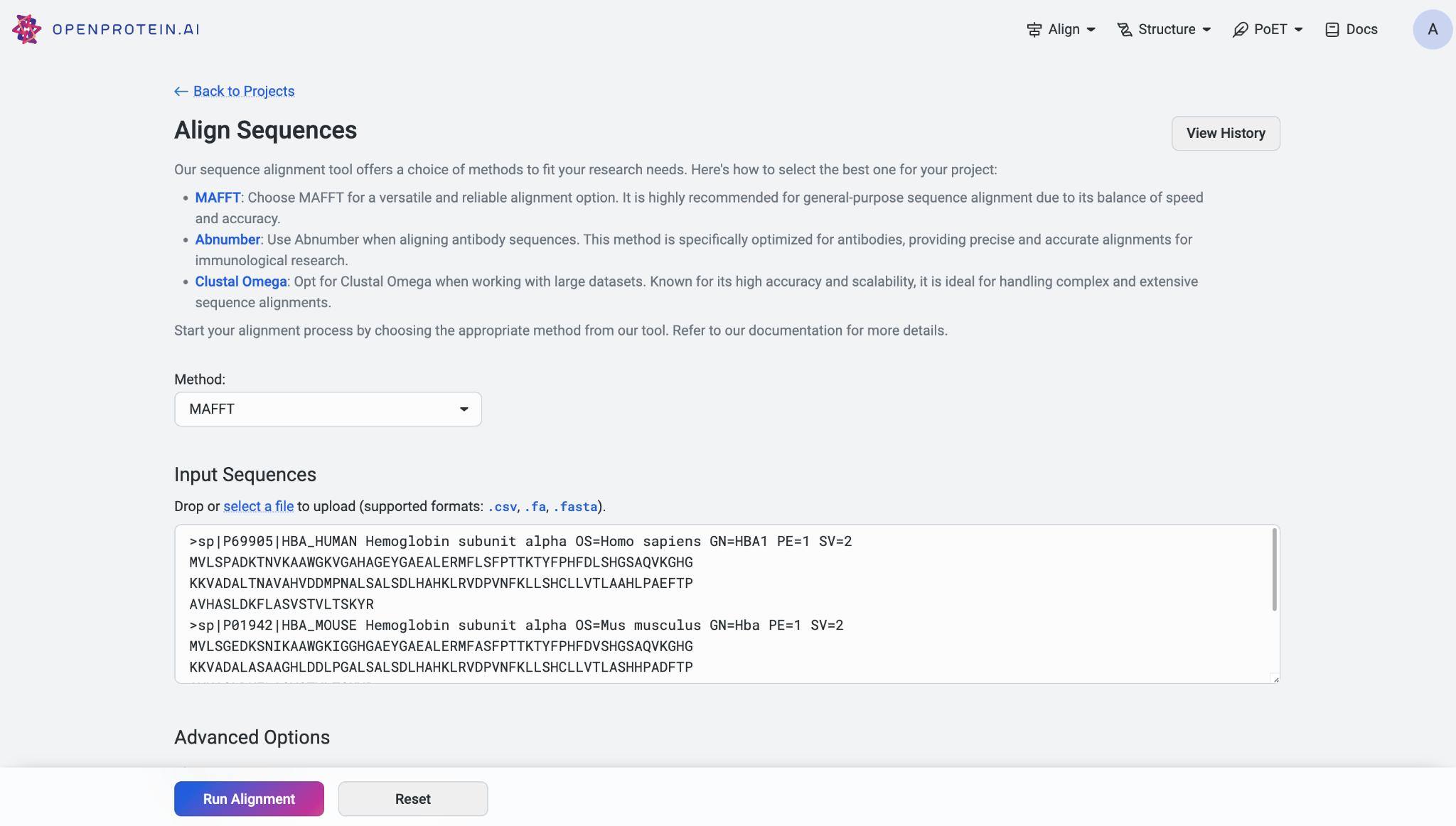
On the alignment page, you have the option to upload your sequence data in various formats, including .csv, .fasta, and .txt. You can either select the upload option to browse and upload a file from your computer or directly copy and paste a list of sequences into the provided input field.
Sequence alignment algorithms#
When preparing to align your sequences, you can choose from different alignment models to suit your needs, including AbNumber, Clustal Omega, and MAFFT. AbNumber works best with antibody sequences, while Clustal Omega is a fast and accurate sequence aligner ideal for large datasets, though it is not suitable for aligning sequences with large internal indels. MAFFT is also a fast and accurate progressive-iterative aligner. MAFFT is the default model.
Customizing your alignment#
Each model offers specific advanced settings to further customize your alignment process. Customizing the parameters can enhance the results, but it is optional; we recommend using the default settings for your first attempt.
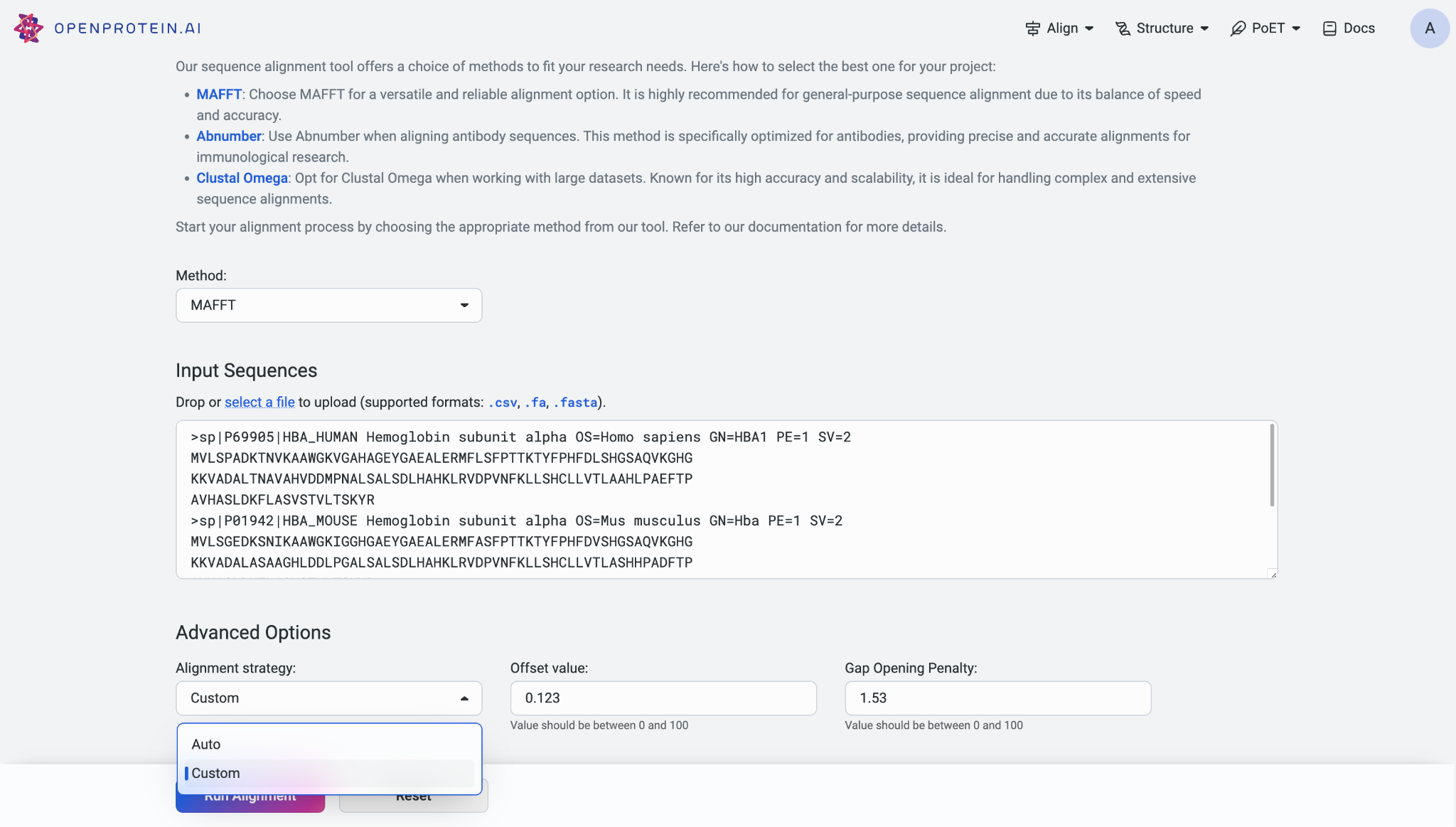
For the AbNumber model, you can select the numbering scheme from the options provided: IMGT (default), Kabat, Chothia, and Aho.
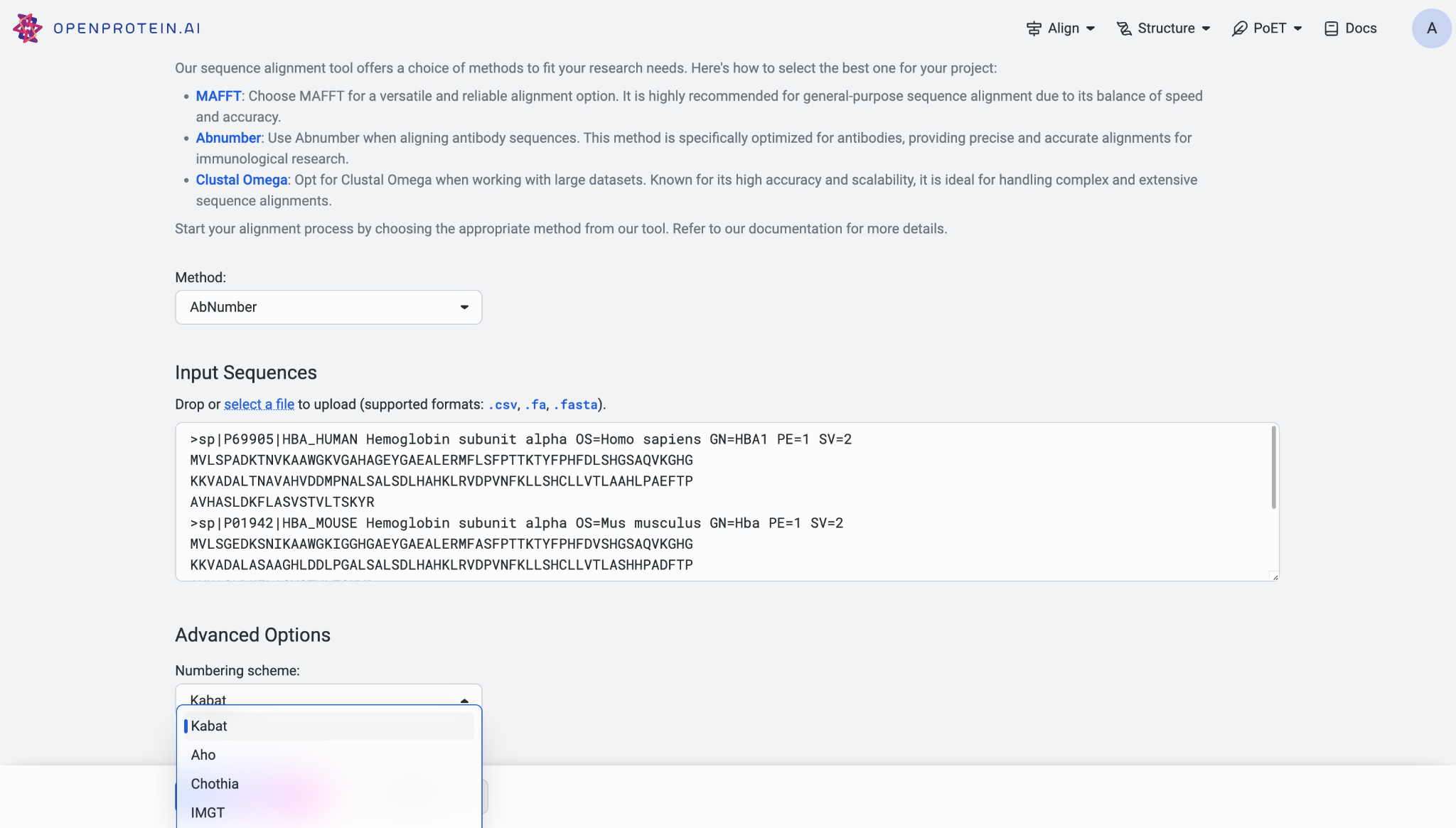
When using Clustal Omega, you can specify the cluster size, with the default set at 100. Sequences are grouped into clusters, and full distance matrices are calculated to construct guide-trees. Additionally, you can define the number of iterations for the fine-tuning process, with a default value of 0.
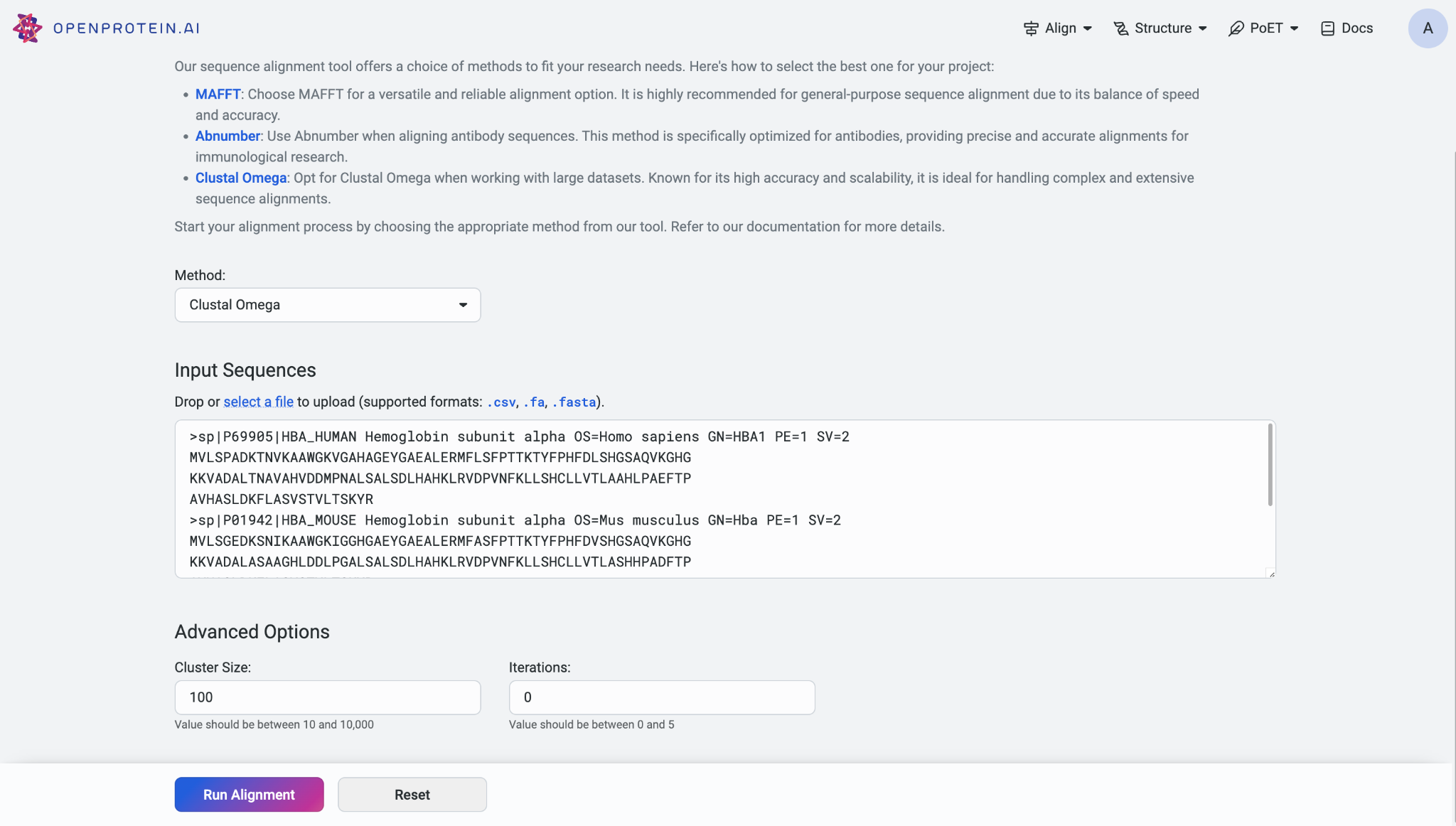
If you opt for the MAFFT model, you can set the alignment strategy to either Auto or Custom. If you select ‘Custom’ as your alignment strategy, you can adjust the offset value, which functions similarly to a gap extension penalty for group-to-group alignment. Additionally, you can set the gap opening penalty, with higher gap penalties useful for closely related matches and lower penalties for identifying more distant matches.
Viewing alignments#
Once the alignment job is completed, the results will be displayed in a table format for easy visualization. You can export the alignment results for further analysis.
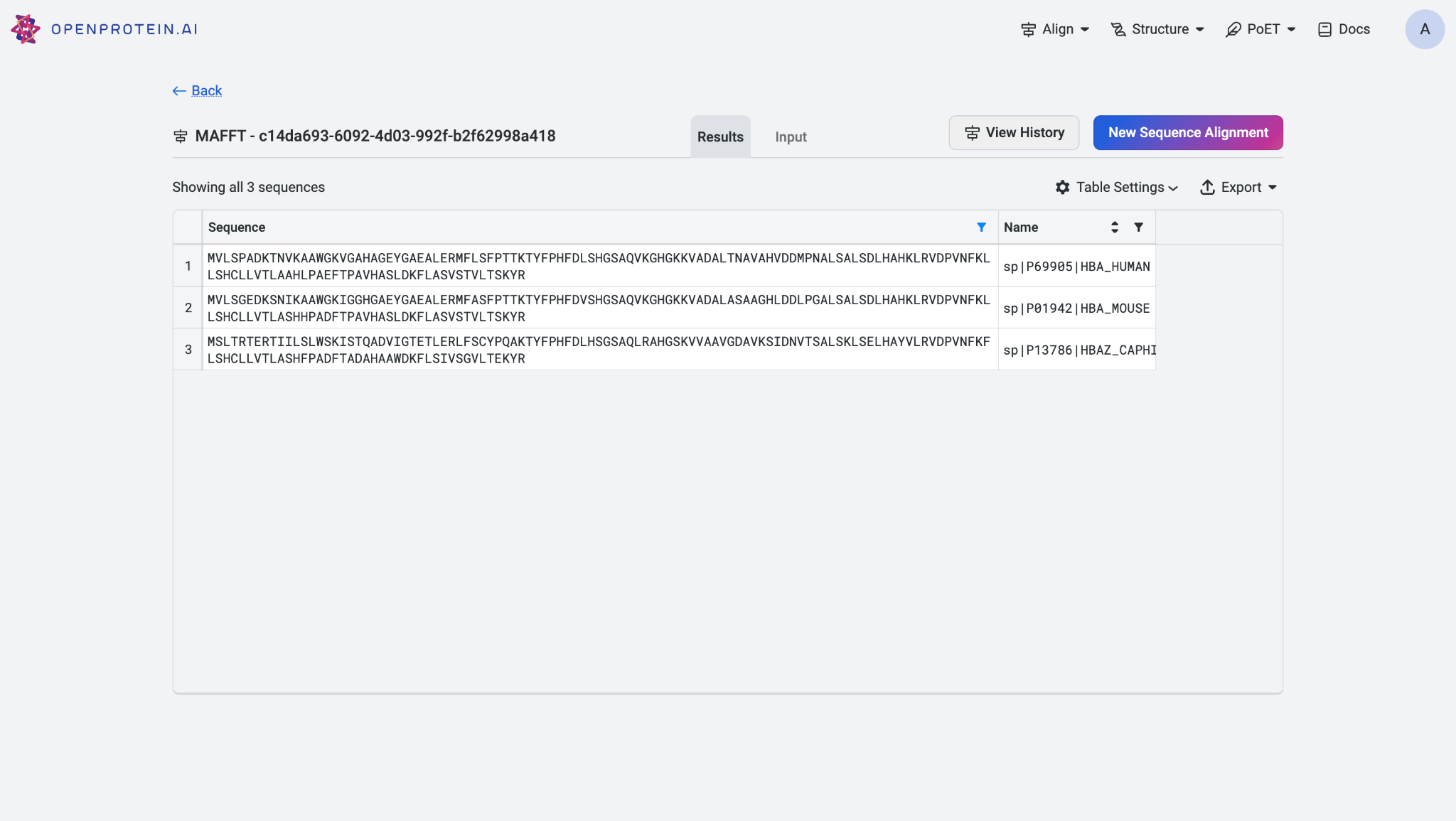
To review the original input sequences and understand the alignment model used, switch to the ‘Input’ tab. To start a new alignment job, click on ‘New Sequence Alignment’.